Data Retrieval
The instructions on this page are for retrieval of Element AVITI and Oxford Nanopore sequencing data only. All other data types (Sanger, TapeStation, Qubit Flex, and Fragment Analysis/GeneScan) are returned through Genomics Depot.
- Using FileZilla on a Windows/Mac/Linux computer
- Using Transmit on a Mac OS X computer
- Using command line wget
- Special instructions for MSU HPCC
- Large data set retrieval
The RTSF Genomics Core FTP server now requires secure (FTPS) connections.
To protect the integrity and security of the MSU network and its systems, and to permit users from outside of the MSU network to once again access the RTSF Genomics Core file distribution server we will be enforcing secure (encrypted) connections only. This change ensures that usernames and password are no longer passed between the client and host in clear text.
For you this will mean using an FTP client application capable of Secure FTP (FTPS) using TLS and configuring it to use an explicit TLS connection. Nearly every current FTP client software is capable of FTPS using TLS. When configuring your software for FTPS use Explicit TLS only, never Implicit TLS. We cannot provide configurations instructions for every FTP client software but below are examples for two popular ones.
FileZilla (Windows/Mac/Linux)
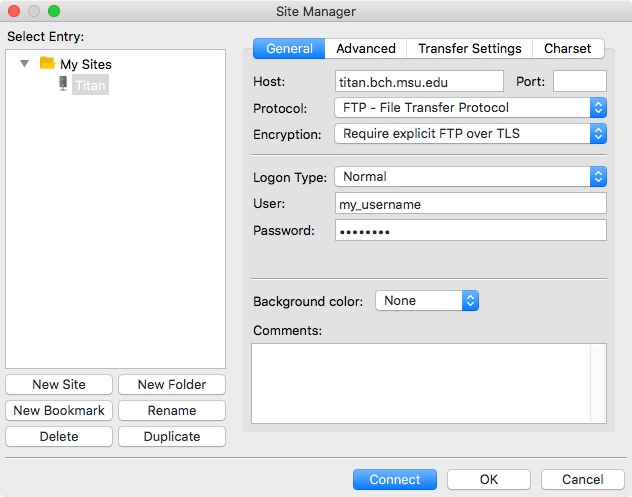
- Open the Site Manager and create a New Site for your RTSF FTPS account.
- Enter the host name titan.bch.msu.edu
- Select Protocol FTP-File Transfer Protocol
- Select Encryption Require explicit FTP over TLS
- Enter the username and password provided to you.
The settings should look like:
Transmit (Mac OS X)
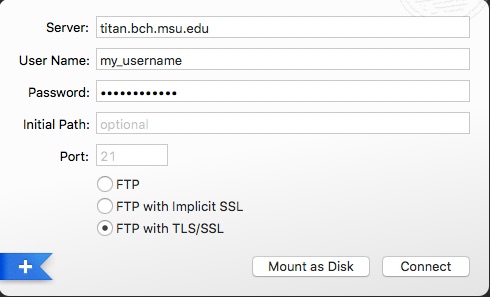
Click on the FTP tab in a new connection window to get the “Connect to FTP Server” dialog.
- Enter the host name titan.bch.msu.edu
- Select the radio button FTP with TLS/SSL
- Enter the username and password provided to you.
The settings should look like:
After clicking Connect with whatever client software you are using you may be presented with a notice that it is unable to verify the server’s certificate. This is normal and you may proceed be clicking OK or Connect, etc.
Transferring data from the RTSF FTP server using the command line program wget
The program wget may be used to transfer data from the RTSF FTP server using the now required encrypted FTP with TLS/SSL (FTPS). There are specific requirements for wget to support FTPS and the wget provided with most standard Linux distributions does NOT support FTPS. Only versions of wget 1.17 or later are capable of supporting FTPS and then only if that support was included when the program was compiled. If you need to install a newer version of wget on your system download the latest version from the GNU Source Repository (http://ftp.gnu.org/gnu/wget/). When running the configure program prior to compiling be sure to include the option "--with-ssl".
If you attempt to run wget as described below and you receive an error message which states: 'Unsupported scheme "ftps"' it means that the version of wget you are using does not support FTPS.
Special considerations for using wget on the MSU HPCC cluster:
You will need to log into the file transfer host (rsync gateway) and load two software modules to perform FTPS file transfers to the HPCC.
rsync.hpcc.msu.edu) will accept SSH keys as the ONLY authentication method. Username/password won't
work. Refer to the ICER SSH key-based authentication Documentation page for instructions to create an SSH keypair and using this to log in to the HPCC.
- log in via ssh using your MSUNetID to rsync.hpcc.msu.edu
$ ssh <MSUNetID>@rsync.hpcc.msu.edu
Do not ssh out to any of the development nodes. FTPS transfer from the RTSF FTP server to the HPCC only works if you are currently working from the rsync login node. Once you have transferred your data to your home directory on the HPCC it will of course be available anywhere on the HPCC.
- load the modules GCCcore v6.4.0 and wget v1.19.4[user@rsyncgw-01]$ module load GCCcore/6.4.0 wget/1.19.4
- execute the wget command[user@rsyncgw-01]$ wget -r -np -nH --ask-password ftps://<username>@<hostname>/<directory_name>
Enter your Genomics FTP account password when prompted.
Username, password and hostname are the ones provided to you (usually via email) when you first received data from the MSU RTSF Genomics Core. directory_name is the name of the subdirectory in you FTP account which contains the data for your current run(s) that you want to download. This subdirectory name is provided in the email notifying you that your data is available.
This command will create a new subdirectory named <directory_name> at your current working directory containing all of the FastQ files for that run.
Large data set retrieval
Oxford Nanopore instruments generate output as both fastq and fast5 formatted files. Typically, you will work with the fastq files which contain your sequence data called with the current basecaller, guppy. These fastq files have been made available to you via FTP download. The sequencer also generates fast5 (HDF5) files which contain the raw signal data from your reads. You may want to rebasecall from fast5 files in the future with more accurate basecalling software. This is unlikely, but it is possible. More importantly, you might want to rebasecall your fast5 files to identify sites of methylation. This is more likely, and thus, the RTSF Genomics Core makes those fast5 files available to you. For PromethION runs, the fast5 files are too large to deliver using the standard FTP server. A run producing 100 Gbp of sequence will have fast5 files totaling 1 TB in size. We will not be able to provide these files via our FTP server, which has limited disk space.
We are no longer backing up data for long term storage to tapes. Fastq files are retained for 1 year. We will store your fast5 files on our network attached storage device for one month, and during that time, you can decide how you may want to receive your fast5 files. After this time your fast5 data may be discarded by us.
These are the options available to you to receive these large fast5 files.
- Globus - Globus is a research data management solution. If you have an existing Globus account simply provide the email address associated with that account and we will send you a link which you may use to transfer your data from our Globus server to a Globus endpoint of your choice. You are responsible for setting up and managing your own Globus account. The RTSF Genomics Core does not provide any technical assistance for this. Resources for MSU researchers may be found on the ICER Documentation Transferring data with Globus page. Researchers outside MSU consult your local support team.
- HPCC - You may identify a directory on HPCC to which you would like your fast5 and fastq files copied. This would preferably be a directory on your scratch space. You must configure the permissions on this directory to make it writable by a designated Genomics Core staff member, or alternatively writable by the world.
- Hard disk - Your data can be copied to a disk drive; MSU researchers can then pick up this drive, off-campus researchers will have it shipped to them. This option costs $85 USD (subject to change, plus shipping) to cover the cost of the drive. This will be a bare, SATA disk drive which you will need to mount inside a computer or external drive case. It will be ext4 formatted which is a native format for Linux. Additional software may be required to read this disk on a Windows or macOS computer. For Windows there is Ext2Read and extFS for macOS.