Sanger Sequencing Sample Requirements
Sanger Sequencing Services to End
Sanger sequencing has been an important service in the Genomics Core since the start of the lab, but during Covid the amount of Sanger submissions to the Genomics Core began to decline. With the rise of plasmid sequencing by Plasmidsaurus and other companies, demand for Sanger sequencing has fallen even further. Prior to Covid, we were sequencing 1,200 to 1,500 sample each week. More recently, we have been receiving 300 to 600 samples per week. This amount of work no longer supports the service contract for our ABI 3730XL. We have made the decision to end Sanger sequencing and fragment analysis services, which also uses the 3730XL. We do not have a precise date for the end of Sanger sequencing. We have sufficient BigDye reagent to allow us to continue sequencing samples until early Spring. We will give further updates when the supply of BigDye is nearly depleted. We will not be hosting a drop-off box for a commercial provider. Several such drop-off sites already exist around campus.
Contact gtsf@msu.edu with any questions.
Overview
The Genomics Core provides a short turnaround time on low- and high-throughput Sanger sequencing projects utilizing the ABI 3730xl. Sequence lengths of 800 to 900 bp are possible (based off PGEM plasmid control).
| Submission Day | Data Available By |
|---|---|
| Monday | Wednesday |
| Tuesday | Thursday |
| Wednesday | Friday |
| Thursday | Monday |
| Friday | Tuesday |
All samples must be submitted with a completed submission form. The submission form and custom naming template can be downloaded from our forms webpage.
On-campus researchers must bring their samples to our hallway drop-off freezer outside of room S-18 in the basement of the Plant Biology Building. Samples should be dropped off between the hours of 8 AM and 4:30 PM Monday through Friday, excluding University holidays.
Samples are accepted from off-campus researchers. Please see our Preparing Samples for Shipping section for details.
Sequence data is returned to researchers through Genomics Depot. If your data is not available after two business days please contact us at gtsf@msu.edu. It is the researcher's responsibility to download and store their data.
Genomics Depot is not for long-term storage and data is only guaranteed for 6 months.
See our Data Retention Policy webpage for more information.
Quick Links
- Sanger Reaction Types
- Service Option Descriptions
- Sample Requirements:
- Preparing Samples for Drop-Off
- Preparing Samples for Shipping
- Where is my data?
- Troubleshooting
Sanger Reaction Types
Standard reaction
- Most common reaction type, will be used by default if selection is not made on submission form.
1st Tier reaction
- For samples with high GC content and plasmids > 10 kb.
- If you have questions about the 1st Tier reaction, please contact the Genomics Core at gtsf@msu.edu.
GTP reaction
- For samples with siRNA hairpins, secondary structure, or if 1st Tier reaction was unsuccessful.
- Please inquire regarding availability and suitability at gtsf@msu.edu.
Researchers can add their own primer to their sample or can request that the Genomics
Core add one of six available in-house primers. Please see the Primer Added by Researcher section for sample requirements when adding your own primer. Please see the Primer Added by Genomics Core section for sample requirements when requesting that the Genomics Core add a primer.
Service Options
The Genomics Core offers four levels of service based on throughput and submission format (i.e. individual 1.7 ml tubes vs. 96-well PCR plate). Pricing for the options below is available on our pricing webpage.
- Samples are submitted in 1.5 ml or 1.7 ml microcentrifuge tubes or 8-strip tubes.
- Individual tubes or 8-strips must be labeled.
- We recommend labeling tubes with your initials and a number (ex. EC1, EC2, EC3, EC4).
- Write legibly on your submission form and tubes. Samples that are unlabeled or illegible will not be processed.
- Below are examples of well labeled 1.5 ml tubes and 8-strip tubes:
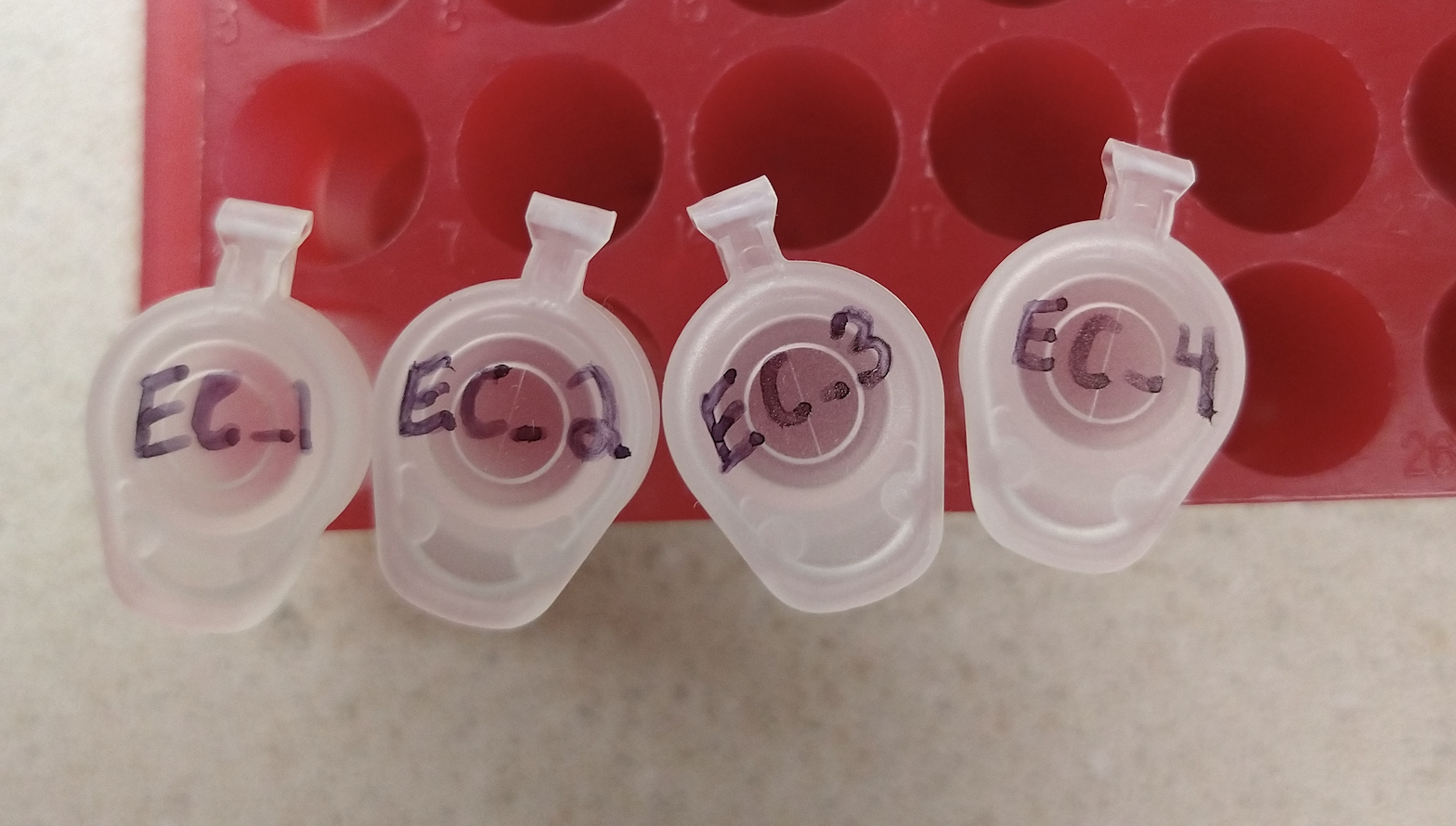

- Complete the top portion of the submission form and provide your sample names in the “Sample names for tube or 8-strips only (maximum 16)” section of the form. View an example of a completed submission form for ≤8 samples in tubes or 8-strips.
- Prepare samples for drop-off and place in submission freezer outside of S-18 Plant Biology or prepare samples for shipping.
- Samples are submitted in 8-strip tubes or 96-well PCR plate.
- For submissions in 8-strip tubes:
- We recommend labeling 8-strip tubes with your initials and number (ex. EC1, EC2, EC3, EC4).
- Write legibly on your submission form and 8-strip tubes. Unlabeled or illegible 8-strip tubes will not be processed.
- Below is an example of well labeled 8-strip tubes:

- Complete the top portion of the submission form and provide your sample names in the “Sample names for tube or 8-strips only (maximum 16)” section. View an example of a completed submission form for 16 samples in 8-strip tubes.
- Prepare samples for drop-off and place in submission freezer outside of S-18 Plant Biology or prepare samples for shipping.
- For submissions in a 96-well PCR plate:
- Use a half-skirt or full skirt 96-well PCR plate and label the side of the plate. Flat-bottom plates will not be accepted.
- Arrange samples by column (vertically) starting with position A1 and do not skip wells. Improperly plated samples will not be processed.
- Write legibly on your submission form and PCR plate. Unlabeled or illegible plates will not be processed.
- Seal the PCR plate.
- Below is an example of a well labeled 96-well plate:

- Complete the top portion of the submission form and the “Sample position and naming for samples in 96-well plate” section. View an example of a completed submission form for 16 samples in a 96-well PCR plate.
- Custom sample names can be used by emailing a custom naming file to gtsf@msu.edu. The custom naming template is available on our forms webpage. Samples will be named by plate name and well position if a naming file is not provided.
- Prepare samples for drop-off and place in submission freezer outside of S-18 Plant Biology or prepare samples for shipping.
- Samples are submitted in a 96-well PCR plate.
- Use a half-skirt or full skirt 96-well PCR plate and label the side of the plate. Flat-bottom plates will not be accepted.
- Arrange samples by column (vertically) starting with position A1 and do not skip wells. Improperly plated samples will not be processed.
- Seal the PCR plate.
- Below is an example of a well labeled 96-well plate:

- Write legibly on your submission form and PCR plate. Unlabeled or illegible plates will not be processed.
- Complete the top portion of the submission form and the “Sample position and naming for samples in 96-well plate” section. View an example of a completed submission form for 40 samples in a 96-well PCR plate.
- Custom sample names can be used by emailing a custom naming file to gtsf@msu.edu. The custom naming template is available on our forms webpage. Samples will be named by plate name and well position if a naming file is not provided.
- Prepare samples for drop-off and place in submission freezer outside of S-18 Plant Biology or prepare samples for shipping.
- *NEW* Low Cost High-Throughput service for 48 or more samples.
- Samples are submitted in a 96-well PCR plate.
- Use a half-skirt of full skirt 96-well PCR plate and label the side of the plate. Flat-bottom plates will not be accepted.
- Arrange samples by column (vertically) starting with position A1 and do not skip wells. Improperly plated samples will not be processed.
- Researcher must add primer.
- NO RERUNS FOR FAILED SAMPLES.
- Seal the PCR plate.
- Below is an example of a well labeled 96-well plate:

- Write legibly on your submission form and PCR plate. Unlabeled or illegible plates will not be processed.
- Complete the top portion of the submission form and the “Sample position and naming for samples in 96-well plate” section. View an example of a completed submission form for NEW low-cost High-Throughput submission in a 96-well plate.
- Custom sample names can be used by emailing a custom naming file to gtsf@msu.edu. The custom naming template is available on our forms webpage. Samples will be named by plate name and well position if a naming file is not provided.
- Prepare samples for drop-off and place in submission freezer outside of S-18 Plant Biology or prepare samples for shipping.
Sample Requirements - Primer Added by Researcher
- Sample concentrations should be measured by fluorometric method (such as Qubit or PicoGreen). UV-based measurements (such as NanoDrop) underestimate sample concentrations and should not be used.
- Sterile water should be used for the sample buffer.
- Add only one primer to each sample. If you would like to sequence with the forward
and reverse primer, two reactions are necessary.
- 30 picomoles of primer should be added. If your primer is at a concentration of 10 µM, you will add 3 µl of your primer to your sample.
- For PCR products, it is very important to purify your sample to remove any remaining primers from the reaction, unincorporated nucleotides, enzyme, buffer, and salts. Make sure any residual ethanol is removed as well. Options for purification include: bead or column-based purification, enzymatic cleanup, or gel extraction.
- If you would like custom sample naming, please download the Sanger custom naming template from the forms webpage. Fill out the template and email it to gtsf@msu.edu before you submit your samples. On your submission form, indicate that custom naming file was emailed to gtsf@msu.edu.
- The volumes requested below are sufficient for two reactions. If less volume is provided, samples cannot be resequenced if the initial reaction fails.
| Sample Type | DNA Mass (ng)* | Volume of 10µM primer to add (µl) |
Total Volume (µl) |
|---|---|---|---|
| Plasmid | |||
| single-stranded DNA | 200 | 3 | 12 |
| double-stranded DNA (up to 10 kb) |
1000 | 3 | 12 |
| Purified PCR Product | |||
| <100 - 200 bp | 2 - 6 | 3 | 12 |
| 200 - 500 bp | 6 - 15 | 3 | 12 |
| 500 - 1000 bp | 10 - 40 | 3 | 12 |
| 1000 - 2000 bp | 20 - 80 | 3 | 12 |
| >2000 bp | 80 - 200 | 3 | 12 |
*These are general guidelines and optimization may be required by researcher.
Sample Requirements - Primer Added by Genomics Core
Researchers can request that the Genomics Core add one of six available in-house primers to their sample (not available for Low Cost High-Throughput service). Before requesting one of the primers below, please verify that your vector has the complimentary priming site.
| Primer Name | Sequence (5' to 3') |
|---|---|
| M13 forward | TGTAAAACGACGGCCAGT |
| M13 reverse | CAGGAAACAGCTATGACC |
| SP6 | ATTTAGGTGACACTATAG |
| T7 | TAATACGACTCACTATAGGG |
| T7 terminator | GCTAGTTATTGCTCAGCGG |
| T3 | ATTAACCCTCACTAAAGGGA |
- Sample concentrations should be measured by fluorometric method (such as Qubit or PicoGreen). UV-based measurements (such as NanoDrop) underestimate sample concentrations and should not be used.
- Sterile water should be used for the sample buffer.
- Do not add a primer.
- If you would like custom sample naming, please download the Sanger custom naming template from the forms webpage. Fill out the template and email it to gtsf@msu.edu before you submit your samples. On your submission form, indicate that custom naming file was emailed to gtsf@msu.edu.
- The volumes requested in the table below are sufficient for two reactions. If less volume is provided, samples cannot be resequenced if the initial reaction fails.
- When requesting that the Genomics Core add a primer to your sample, you must indicate on your submission form which primer is needed. If you will request more than one primer, you must group your samples together by column that will receive the same primer. Examples of how to arrange samples when the Genomics Core will add the primer are below. The left image is for submissions in individual tubes or 8-strip tubes and the right image is for submissions in a 96-well PCR plate.
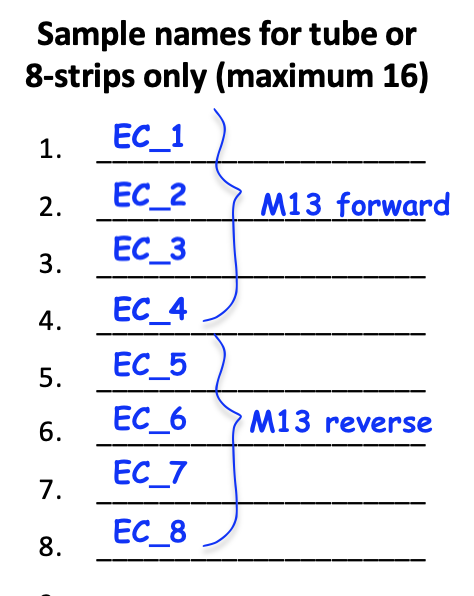
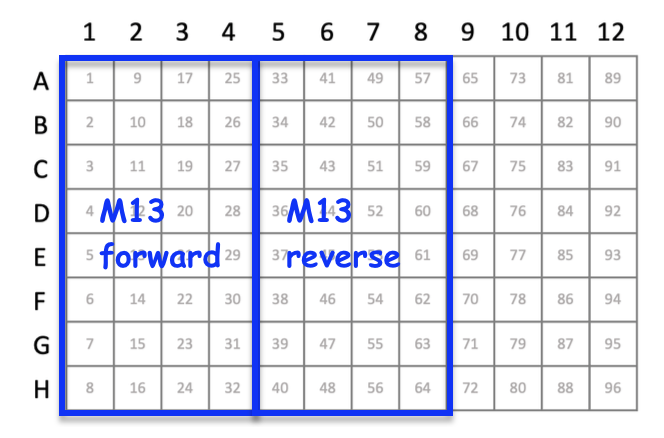
| Sample Type | DNA Mass (ng)* | Total Volume (µl) |
|---|---|---|
| Plasmid | ||
| single-stranded DNA | 200 | 8 |
| double-stranded DNA (up to 10 kb) |
1000 | 8 |
| Purified PCR Product | ||
| <100 - 200 bp | 2 - 6 | 8 |
| 200 - 500 bp | 6 - 15 | 8 |
| 500 - 1000 bp | 10 - 40 | 8 |
| 1000 - 2000 bp | 20 - 80 | 8 |
| >2000 bp | 80 - 200 | 8 |
*These are general guidelines and optimization may be required by researcher.
Preparing Samples for Drop-Off
Prepare samples in the appropriate format and complete the submission form as described in the Service Options and Sample Requirements sections above.
Individual tubes or 8-strip tubes should be put in a zip-lock bag, rack, or box. Plates can be placed in a zip-lock bag or box if desired, but it is not necessary. Zip-lock bags, racks, and boxes are available outside of the submission freezer if needed. Submission forms are also available outside of S-18 Plant Biology.
Label the bag, rack, or box with your name, your PI/Lab name, and the date. Examples of properly labeled secondary containers are below:

Bring samples (in bag, rack, or box) to S-18 Plant Biology and place them in the submission freezer in the box labeled "Sanger Sequencing Submissions".

Place the submission form in the "Sanger Submissions" basket hanging on the outside of freezer.

Email gtsf@msu.edu with any questions.
Preparing Samples for Shipping
Prepare samples in the appropriate format and complete the submission form as described in the Service Options and Sample Requirements sections above.
Off-campus researchers should be aware that “overnight deliveries” from both FedEx and UPS are often taking two days. These overnight packages are being delayed in warehouses outside of Michigan. Once packages arrive in East Lansing, they are being delivered correctly, and we are having no problems with packages that make it to our building. Because of the probability of delay, samples should be sent with a gross excess of dry ice. Additionally, plastic film seals for 96-well PCR plates should be avoided as thawed samples readily leak into adjacent wells. To avoid leakage, we recommend an excess of dry ice and using strip caps that were designed for the 96-well PCR plate or sealing foil. We do not recommend shipping at ambient temperature or on wet ice. USPS is not recommended for shipments.
When shipping on dry ice, do not place 96-well PCR plates or tubes directly into the dry ice. Doing so can result in broken or cracked plates/tubes and loss of samples. Place samples in a secondary container that is rigid, such as a pasteboard freezer box (tip boxes may also work well) that is taped closed. The secondary container can then be placed in dry ice. If using strip caps, please tape them down.
Address shipments to:
Colleen Curry
612 Wilson Road, Rm S-18 D
East Lansing, MI 48824
Email carrier and tracking information to gtsf@msu.edu so that we know to expect your package.
Where is my data?
Sequence data is returned to researchers through Genomics Depot. If your data is not available after two business days please contact us at gtsf@msu.edu. It is the researcher's responsibility to download and store their data. Genomics Depot is not for long-term storage and data is only guaranteed for 6 months. See our Data Retention Policy webpage for more information.
Sequence data files are ab1 file format. Researchers will need software to view and analyze this file type. There are many software options available. Some options to explore are: Sequence Scanner, SnapGene, 4Peaks, Chromas Lite, Chromas Pro, GeneStudio Pro, UGENE, Chromaseq, Geneious, SeqMan Pro, Sequencer, Vector NTI Express, CodonCode Aligner, DNA Baser, BioLign, CAP3, ApE, and EveryVector. Please note that the software mentioned here may be free or be available for a fee. This is not an all-inclusive list.
Troubleshooting
The RTSF Genomics Core has developed a Sanger Sequencing Guide to demonstrate best and worst practices when performing Sanger Sequencing. This guide shows examples of good- and poor-quality sequence to help researchers to troubleshoot their own Sanger Sequencing results. The guide and the files used in the guide can be downloaded from the Sanger Sequencing Best and Worst Practices webpage. If you need more assistance troubleshooting your results, please contact us at gtsf@msu.edu.