Amplicon/Metagenomic Guide
Overview
This webpage provides details, recommendations, and requirements for the amplicon library preparation services offered by the MSU Genomics Core. The table below provides an overview of the submission requirements for 16S-V4 and all other amplicons (for barcoding). Additional requirements, PCR recipes, and cycling conditions are provided below the table.
The Genomics Core will include a negative, no template control (NTC) reaction on each plate of samples processed for amplicon sequencing, both 16S and other amplicons. Researchers must leave at least one well empty (H:12) on each plate submitted for amplicon library preparation. The NTC added by the Genomics Core substitutes 1 µl of nuclease-free water for the usual 1 µl of template DNA in the PCR reaction. This reaction is processed and sequenced alongside your samples. The purpose for including this negative control is to demonstrate that good laboratory practices are being followed in the core facility, that no contamination of reagents or cross contamination of samples has occurred. Researchers are encouraged to include their own negative controls to monitor contamination introduced by sampling, DNA extraction and primary PCR reactions prior to sample submission to the Genomics Core.
For 16S-V4 amplicon projects, the Genomics Core offers the option of including a positive control in your project, the ZymoBIOMICS Microbial Community DNA Standard (Zymo Research Cat# D6305/D6306). This mock community is composed of defined proportions of genomic DNA from eight bacteria plus two fungi, and sequencing of this mock community can serve as a useful positive control. Addition of a single positive control sample will be at no additional charge.
16S-V4 Amplicons
Amplicon Barcoding
References
| Target | Input | Concentration | Minimum Volume | Multiplex | Notes |
|---|---|---|---|---|---|
| 16S-V4 | Metagenomic DNA | 1 ng/µl minimum | 10 µl | 24 to 380 samples (unique dual indexes) | Test amplification and gel image required, see below for more details. |
| Amplicon Barcoding | Primary PCR product | 1 ng/µl minimum | 10 µl | 24 to 380 samples (unique dual indexes) | Gel image required, see below for more details. |
16S-V4 Amplicons
The library preparation for the 16S-V4 region employs a one-step PCR method using the primer pair 515f/806r:
16S V4 forward (515f): GTGCCAGCMGCCGCGGTAA
16S V4 reverse (806r): GGACTACHVGGGTWTCTAAT
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K. & Schloss, P. D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Applied and environmental microbiology 79, 5112–5120 (2013) (https://github.com/SchlossLab/MiSeq_WetLab_SOP/blob/master/MiSeq_WetLab_SOP.md)
The submitter is required to:
- Quantify samples via fluorometric method (such as Qubit or PicoGreen) and normalize all samples to approximately the same concentration.
- Perform a test amplification using the 16S-V4 primers (above) to confirm the sample can be amplified and that the product is the expected size. A ladder must be run alongside the samples so that the PCR products can be sized. The gel must be run long enough to have good separation of the ladder. The gel image(s) must be provided at the time of submission.
- Submit samples in a 96-well PCR-type plate, filled by column, starting with A:1. Well H:12 must be left empty for the RTSF no template negative control.
The Genomics Core will use 1 µl of genomic DNA for the amplicon library preparation. Therefore, it is recommended that test amplifications be performed using a protocol similar to the Genomics Core's amplicon library preparation protocol:
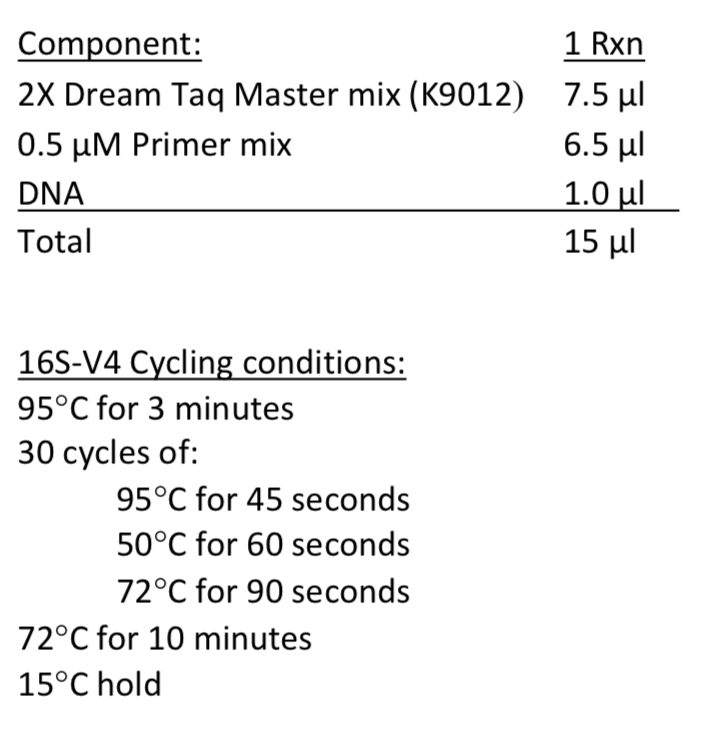
AMPLICON BARCODING (INDEX ADAPTERS ADDED BY MSU GENOMICS CORE)
For all other amplicon sequencing, a two-step PCR approach is used. The first PCR uses target-specific primers with tags on the 5’ ends that allow the Genomics Core to do a second PCR for barcoding. The submitter is required to perform the primary PCR reaction. To do this, the submitter will need to order primers with the following tags on the 5’ ends, with the target-specific forward and reverse sequences inserted in [TS-For] and [TS-Rev], respectively:
CS1-TS-F: 5’ – ACACTGACGACATGGTTCTACA – [TS-For] – 3’
CS2-TS-R: 5’ – TACGGTAGCAGAGACTTGGTCT – [TS-Rev] – 3’
When choosing primers for a targeted amplicon it is recommended that the product amplified is less than 500bp (not including the 44bp added by the CS1/CS2 overhangs). This size will permit adequate overlap of the forward and reverse read 3' ends when sequencing with a 2x300bp format.
The Genomics Core will use 1 µl of primary PCR product for the amplicon indexing.
The submitter is required to:
- Quantify samples via fluorometric method (such as Qubit or PicoGreen) and normalize all samples to approximately the same concentration.
- Run products on a gel to confirm the amplification was successful, the products are the expected size, and that off-target products (including primer-dimer) are not present. The gel image must be provided at the time of submission.
- Products do not need to be cleaned up prior to submission. However, if primer dimers or off-target products are generated, then products should be cleaned up to remove these products and/or the primary PCR reaction should be optimized.
- Submit samples in a 96-well PCR-type plate, filled by column, starting with A:1. Well H:12 must be left empty for the RTSF no template negative control.
References
Kozich, J. J. et al. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Applied and environmental microbiology 79, 5112–5120 (2013).
Klindworth, A. et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41, e1–e1 (2013).