Fall 2024 Event Schedule
The MSU Genomics Core has scheduled the following events for Fall 2024. Click on the titles below to jump to a full description of the event.
October 1st - Designing Single-Cell RNA-seq Experiments Lecture
October 9th - AVITI Introduction Seminar
October 17th - Genomics Core Open House
Designing Single-Cell RNA-seq Experiments Lecture
Tuesday, October 1st, 10 AM
Food Safety and Toxicology Building, Room 162
Stephanie Lepp Hickey, Ph.D., Associate Director of the MSU Bioinformatics Core
Single-cell RNA sequencing (scRNA-seq) has transformed our understanding of cellular diversity by offering detailed insights into gene expression at the single-cell level. However, the complexity of scRNA-seq demands careful experimental design to ensure reliable and reproducible results. This lecture will cover the essential aspects of designing scRNA-seq studies, including setting clear research objectives, and determining the right number of cells and replicates. We will also address technical considerations like cell isolation, library preparation, and sequencing depth, and how these affect data quality. By the end of the session, participants will understand the key principles of scRNA-seq experimental design, enabling them to conduct studies that yield high-quality, impactful data.
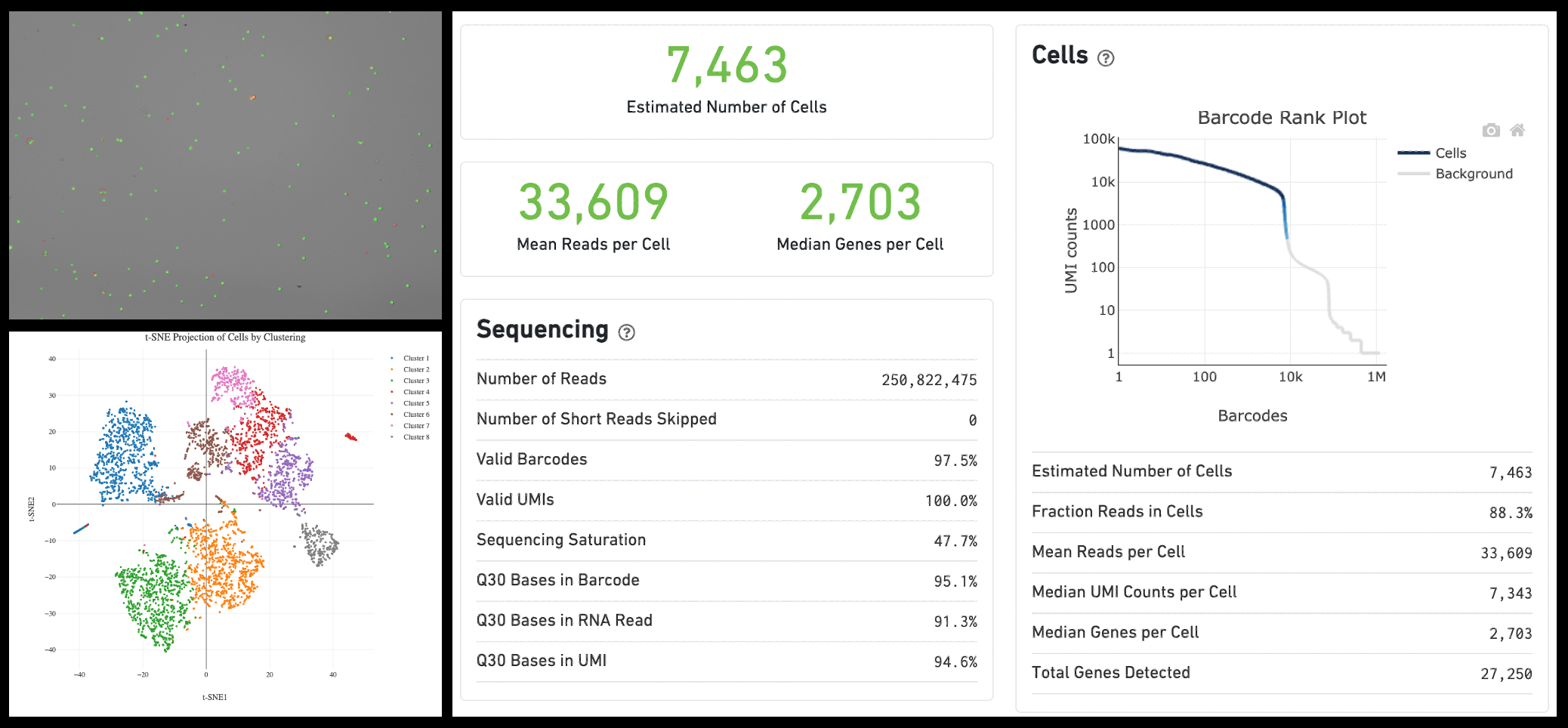
AVITI Introduction Seminar
Missed the seminar? View and download the AVITI Introduction Seminar Slides.
Wednesday, October 9th, 12 PM (noon)
Interdisciplinary Science and Technology Building, Room 1404
Solomon Hailu, Ph.D, Element Biosciences
Kevin Childs, Ph.D, Director of the MSU Genomics Core
The MSU Genomics Core has acquired an AVITI sequencing instrument from Element Biosciences. This instrument will replace our NovaSeq 6000, which has a planned retirement date of late December. The seminar will discuss details about how the AVITI will change sequencing in the MSU Genomics Core, and it will discuss Avidity-Base Chemistry (ABC) sequencing technology. The seminar is being hosted by Element Biosciences and the MSU Genomics Core.
Briefly, the AVITI uses Avidity-Base Chemistry (ABC) sequencing as opposed to Illumina’s Sequencing By Synthesis (SBS) technology. The AVITI has a higher accuracy, lower duplication rate, and no index hopping compared to the NovaSeq 6000. Average read quality is Q30 for 93% of reads and Q40 for >80% of reads. Read duplication rates are ~2% compared to more than 20% read duplication with the NovaSeq. Standard short read libraries that researchers have been using for years can be run on the AVITI with no modifications. The AVITI outputs the same fastq files that researchers expect, and no changes will be necessary in data analysis pipelines. Importantly, flow cells on the AVITI generate 250, 500 and 1,000 million read pairs, which means that sequencing projects will have a faster turn-around time than with the NovaSeq. Finally, sequencing costs will be significantly cheaper with the AVITI than with the NovaSeq 6000.
More information about the AVITI System is given below the flyer.
Lunch will be provided. Please registered at the link below.
https://go.elementbiosciences.com/MSUseminar
Introducing the Element AVITI Sequencing System
The Element AVITI System reimagines the core components of NGS to offer a benchtop platform that grants access to the genomics ecosystem. Delivering flexible throughput at exceptionally low cost, AVITI saves time and resources without the need to batch samples. Avidity base chemistry (ABC) forms the core of a disruptive design that readily adapts to any application, offering methods that scale from amplicon to whole genomes.
Novel ABC Sequencing
The fundamentals of ABC technology leverage the unique properties of avidites to execute an efficient sequencing reaction and yield highly accurate data. A strong signal-to-noise ratio that persists through high polony densities drives this accuracy. When a run starts, the library hybridizes to surface primers coating the flow cell. Amplification polymerase then binds to the library and primer duplexes, catalyzing rolling circle amplification (RCA) and generating long DNA strands that include copies of the original library. Each strand forms a polony that contains hundreds of copies of the original library. The polonies hybridize to read-specific sequencing primers. For each cycle, a sequencing polymerase binds an avidite to a polony and primer duplex and traps a base-specific avidite to the polony. The result forms an extremely tight complex that enables a 100-fold reduction in reagent concentration compared to sequencing-by-synthesis chemistry (SBS). AVITI and ABC reset expectations on quality scores (Q scores), delivering exceptional Q30 accuracy for 2 x 150 sequencing at > 90% and > 85% for 2 x 300 sequencing. AVITI demonstrated higher accuracy compared to legacy sequencing technology. Data showed fewer soft-clipped reads in difficult homopolymer and repeat regions, among other clear advantages. AVITI is directly compatible with most linear third-party libraries.
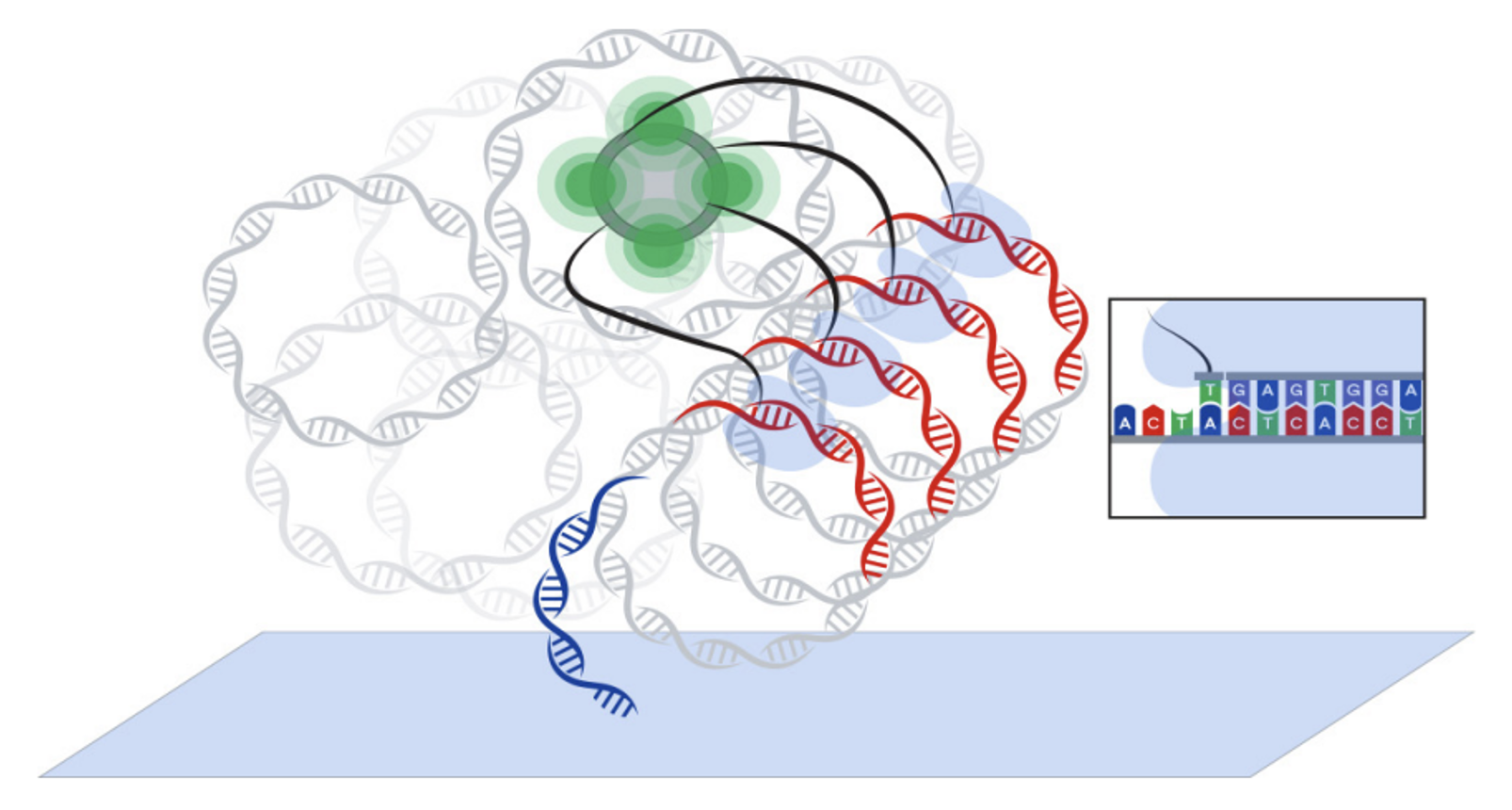 Polymerase binds avidites at the interrogation site. The avidite arms connect to a
core that provides a fluorescent signal for detection.
Polymerase binds avidites at the interrogation site. The avidite arms connect to a
core that provides a fluorescent signal for detection.
Download the infographic to learn more about Avidite Base Chemistry.
Highest accuracy sequencing
Standard Cloudbreak sequencing kits consistently generate greater than 90% bases at Q40 (estimated error rates 1 in 10,000).
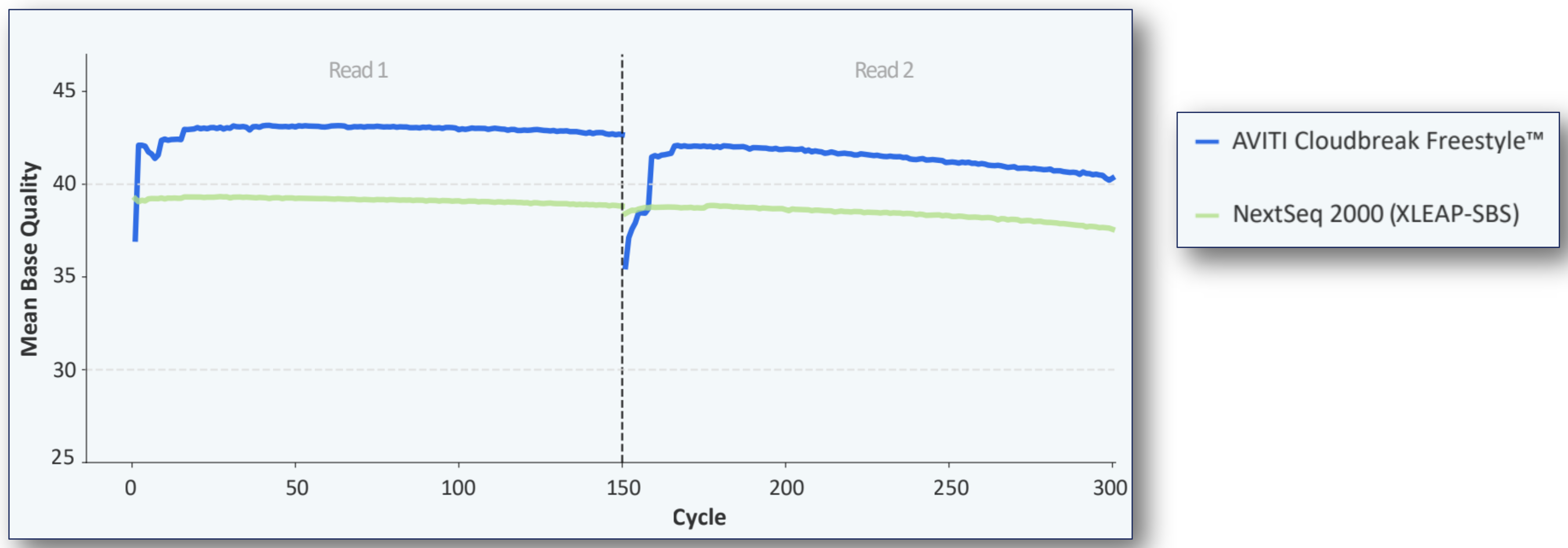
Base quality of AVITI Cloudbreak Freestyle and NextSeq 2000 (XLEAP-SBS) 2x150 sequencing
Flexible read lengths and throughput
Sequencing kits for the AVITI platform range from 2x75 bp paired end, ideal for tag-counting applications such as RNA-seq, to 2x300 bp paired end kits for applications that require longer reads including amplicon sequencing. Low, medium, and high output flow cells are available to adjust data output. By processing two independent flow cells, multiple configurations can run simultaneously to enable faster turnaround times.
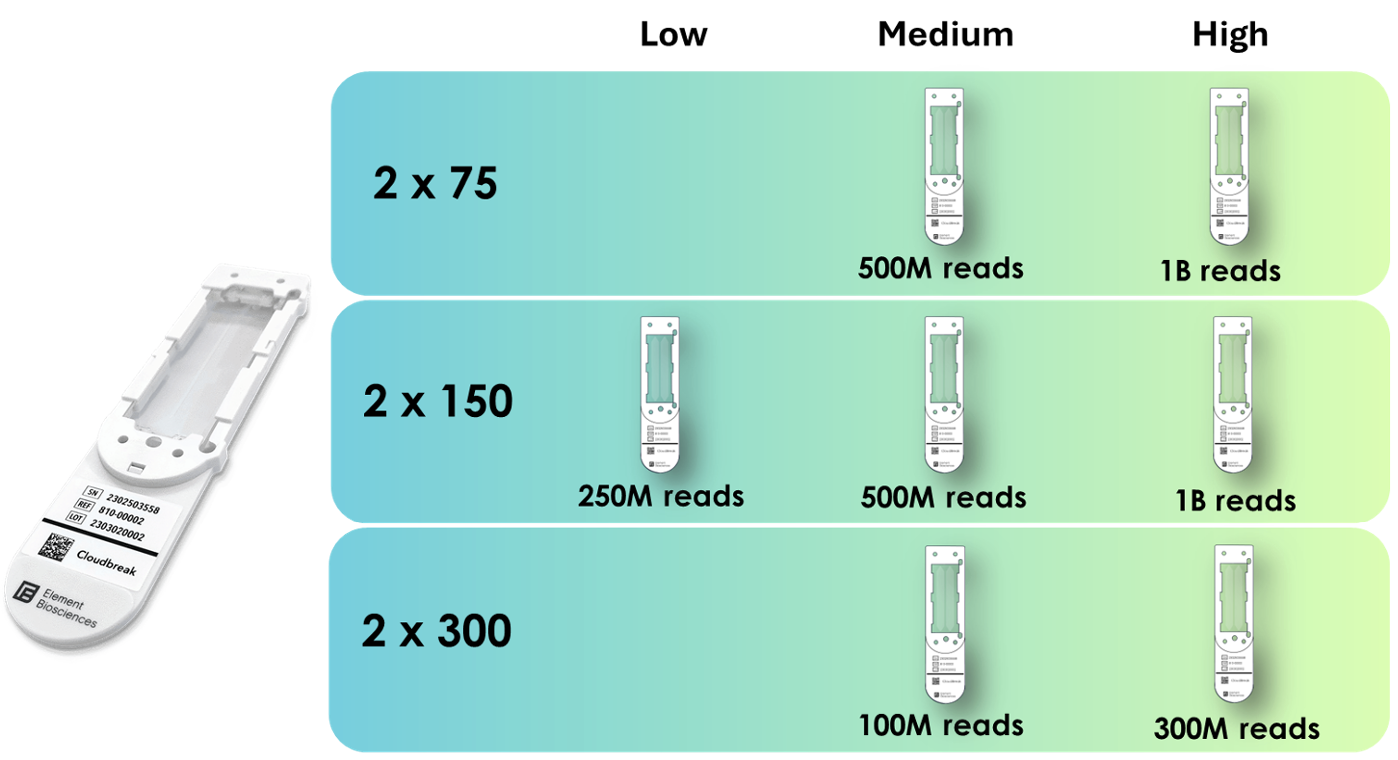
Flow cell configurations available at the MSU Genomics Core.
Genomics Core Open House
Thursday, October 17th, 1 - 4 PM
Plant Biology Building, Room S-18

